Bioimage analysis is a crucial process in modern biological research, focusing on extracting quantitative data from biological images. This analysis enables researchers to gain deeper insights into cellular and tissue-level phenomena, providing critical information for various applications in medicine, pathology, and biomedical sciences. Due to the inherent complexity of biological images, sophisticated algorithms from fields such as image processing and artificial intelligence (AI) play an essential role in ensuring accurate and efficient analysis. These algorithms enable tasks like segmenting complex structures, detecting subtle patterns, and classifying features within the images with high precision, all of which are vital to understanding the underlying biological systems.
In addition to traditional image processing techniques, machine learning models further enhance bioimage analysis by learning from existing data and adapting to the unique conditions of diverse imaging modalities. These models can improve the accuracy of analysis over time, handling a wide range of variability within biological data. As a result, these computational approaches allow researchers to analyze vast datasets both rapidly and with great precision, driving significant advancements in fields like pathology, cancer research, and drug discovery.
This document presents a case study focused on data fusion from two digital pathology tools, one selected from a list of free tools and the other from a set of commercial tools. By combining the strengths of these two tools, the case study demonstrates how using different types of software can enhance bioimage analysis workflows and improve overall efficiency.
Choice of tools
Several free image analysis tools are available for use in digital pathology, including Icy, ImageJ, CellProfiler, Napari, and QuPath. Among these, QuPath stands out due to its intuitive graphical user interface (GUI), fast machine learning capabilities, and seamless integration with other platforms. QuPath’s flexibility allows it to work in tandem with ImageJ and Python-based libraries, enabling users to extend its core functionality for specific research needs. One of QuPath’s key strengths is its ability to handle very large whole slide images (WSIs), which are often difficult for other tools to manage. Its zooming capacity is another significant advantage, allowing users to annotate intricate, fine details with ease. For these reasons, QuPath was chosen as the free tool for the analysis discussed in this case study.
On the other hand, commercial tools for digital pathology offer certain advantages, including a more polished and stable environment, as well as enhanced support and user experience. Prominent tools in this category include MIPAR, Visiopharm, HALO, PathAI, and Biodock. While many of these platforms excel in providing AI-driven analysis for 2D pathology images, Biodock was selected for its unique combination of cost-effectiveness, simplicity, and cloud-native infrastructure. Biodock’s cloud-based design allows users to access and analyze data from any location, making it particularly useful for collaborative research efforts. Its scalability enables users to adjust processing power based on the volume and complexity of their data, ensuring efficient performance even with large datasets. Additionally, Biodock offers a marketplace where users can access pre-trained models developed by leading researchers and institutions. Biodock’s ease of use, flexibility, and cloud-based scalability made it the preferred choice among the commercial tools considered for this study.
By combining the strengths of QuPath and Biodock, this case study highlights the potential benefits of using both free and commercial digital pathology tools in bioimage analysis. The free tool provided flexibility and ease of integration, while the commercial tool offered a stable, scalable, and user-friendly platform. Together, these tools facilitated a comprehensive and efficient analysis, showcasing the ways in which digital pathology software is driving advancements in biological research.
Data
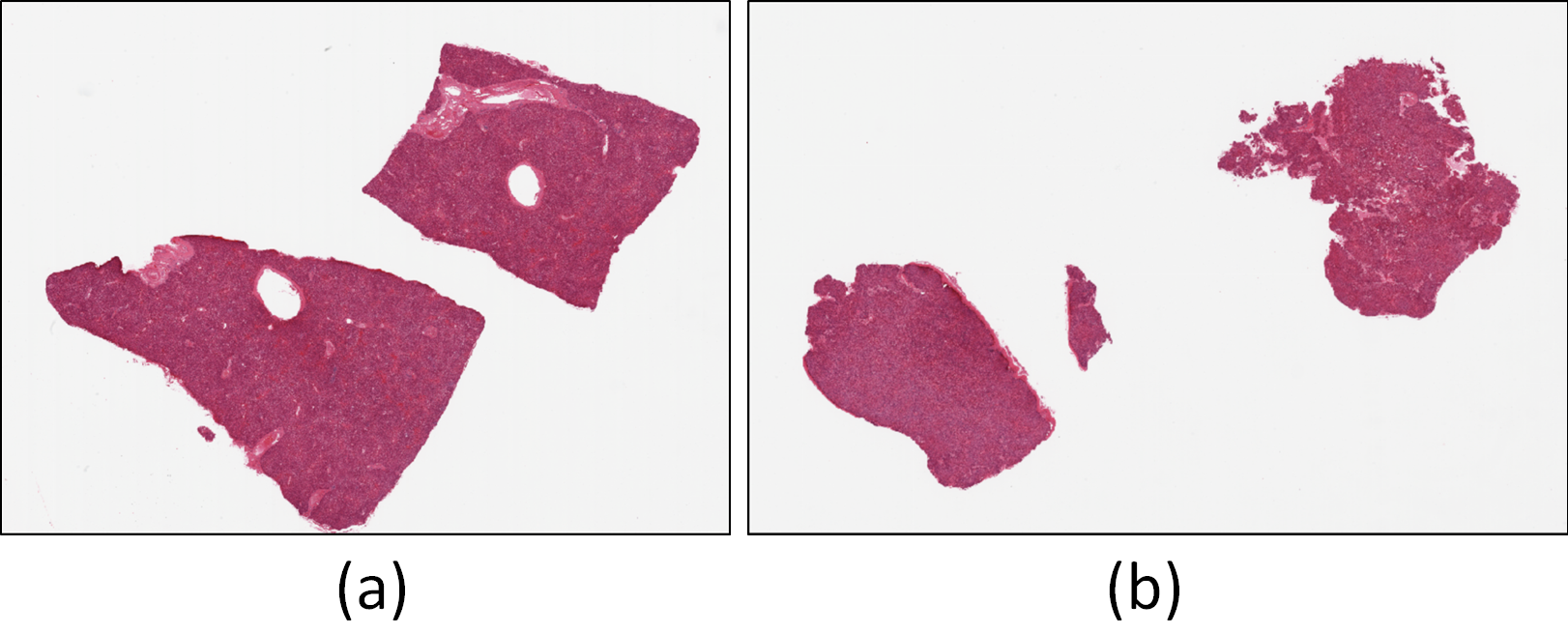
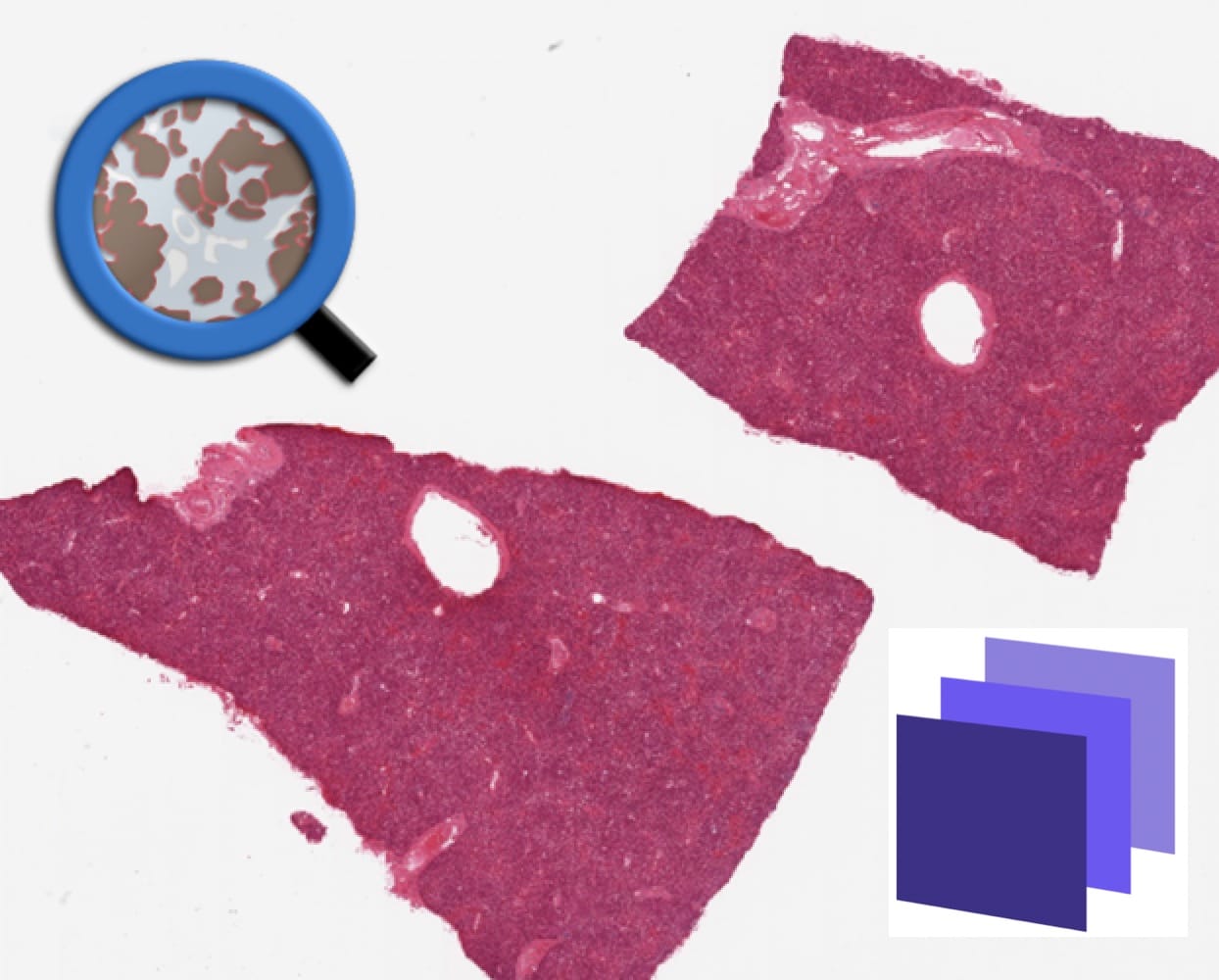
The data for this experiment was sourced from the GTEx portal (https://gtexportal.org/home/histologyPage), a well-established resource for histological and genetic data. Two whole slide images of H&E stained liver tissues, corresponding to the sample IDs GTEX-11GSP-0626 (Fig. 1(a)) and GTEX-11NUK-1226 (Fig. 1(b)), were selected for use in both training and testing machine learning models within QuPath and Biodock. Processing whole slide images, however, is highly demanding. Not only does it require a system with significant computational resources, but the time needed to analyze and process the images can also be quite substantial. To mitigate this challenge, and to improve efficiency without compromising the quality of the data, twelve smaller sections were carefully extracted from the whole slide images. Specifically, seven sections were taken from GTEX-11GSP-0626 and five from GTEX-11NUK-1226. Each section was extracted with a resolution of 6000 x 6000 pixels, which strikes a balance: it reduces the processing time while ensuring that crucial histological details necessary for accurate training are still captured. This resolution was chosen specifically to avoid missing key information during the model training phase. For the sectioning process, the 'openslide' library for Python 3 was employed, a powerful tool that facilitates the manipulation and extraction of smaller image regions from large, complex whole slide images.
Process of analyses
The expectation from the analyses were the identification of nuclei, portal tracts, and the nuclei within each portal tracts in the liver tissue.
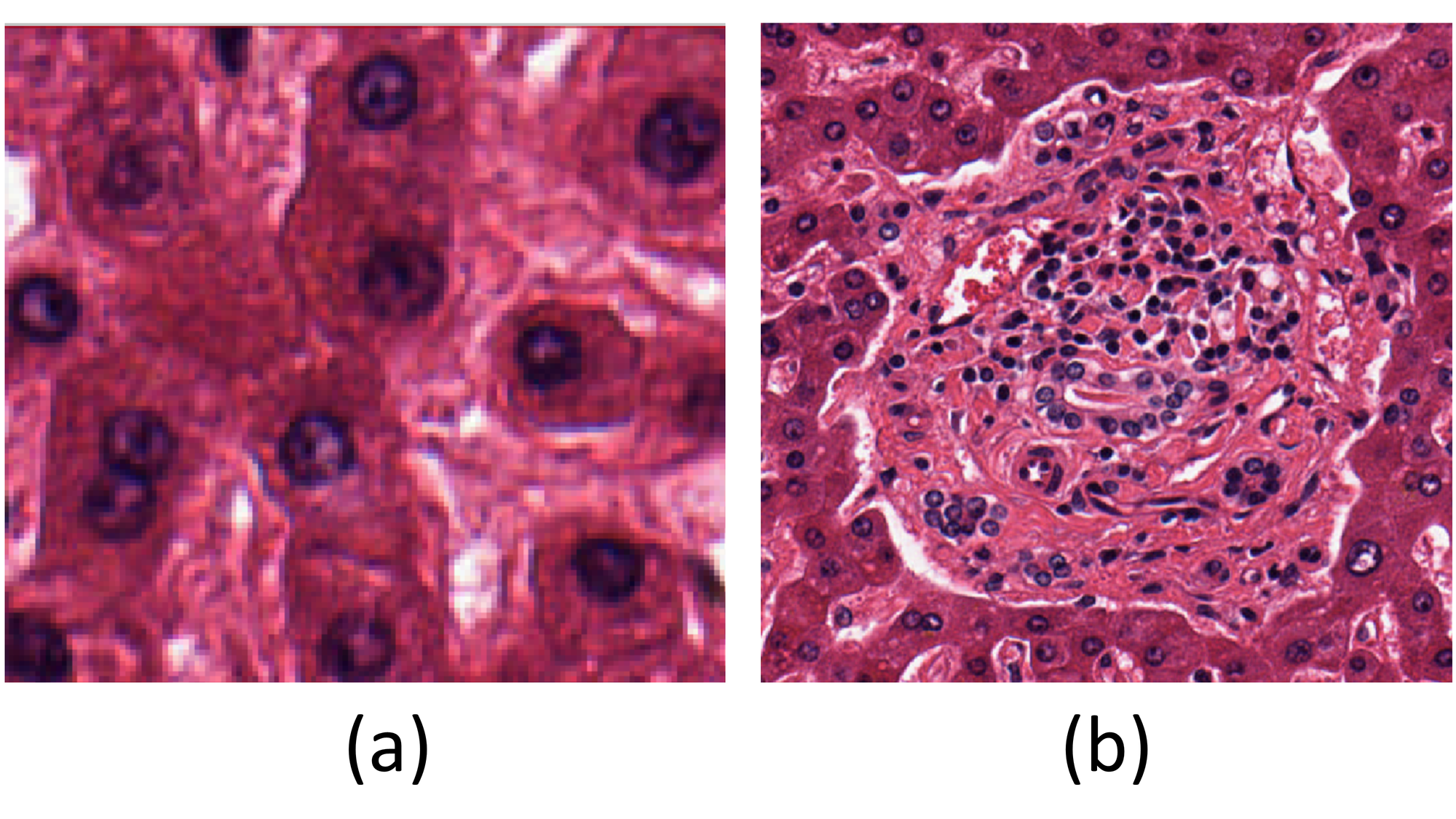
As previously discussed, one task of analysis was assigned to QuPath, which runs on a local machine, so a pattern requiring less computational power was chosen for it. The liver tissue images considered for the case study contain a high concentration of nuclei, making nuclei detection (Fig. 2(a)) an ideal choice for QuPath. This tool offers a cell detection option where properties can be adjusted to identify enclosed objects like nuclei.
On the other hand, the task assigned to Biodock was to identify portal tracts in the liver tissue images. A portal tract (Fig. 2(b)) includes branches of the portal vein, hepatic artery, bile ducts, lymphatic vessels, nerves, and connective tissue, making it a complex structure to detect due to its intricate pattern, computational demands, and time requirements. However, Biodock’s cloud-native infrastructure alleviated the computational burden on the local machine.
Since the nuclei within each portal tract also needed to be obtained, the workflow was initiated by performing the analysis on Biodock first, followed by downloading the portal tract masks for further analysis in QuPath. The steps taken for training and testing in Biodock are shown in Fig. 3 and Fig. 4, respectively.
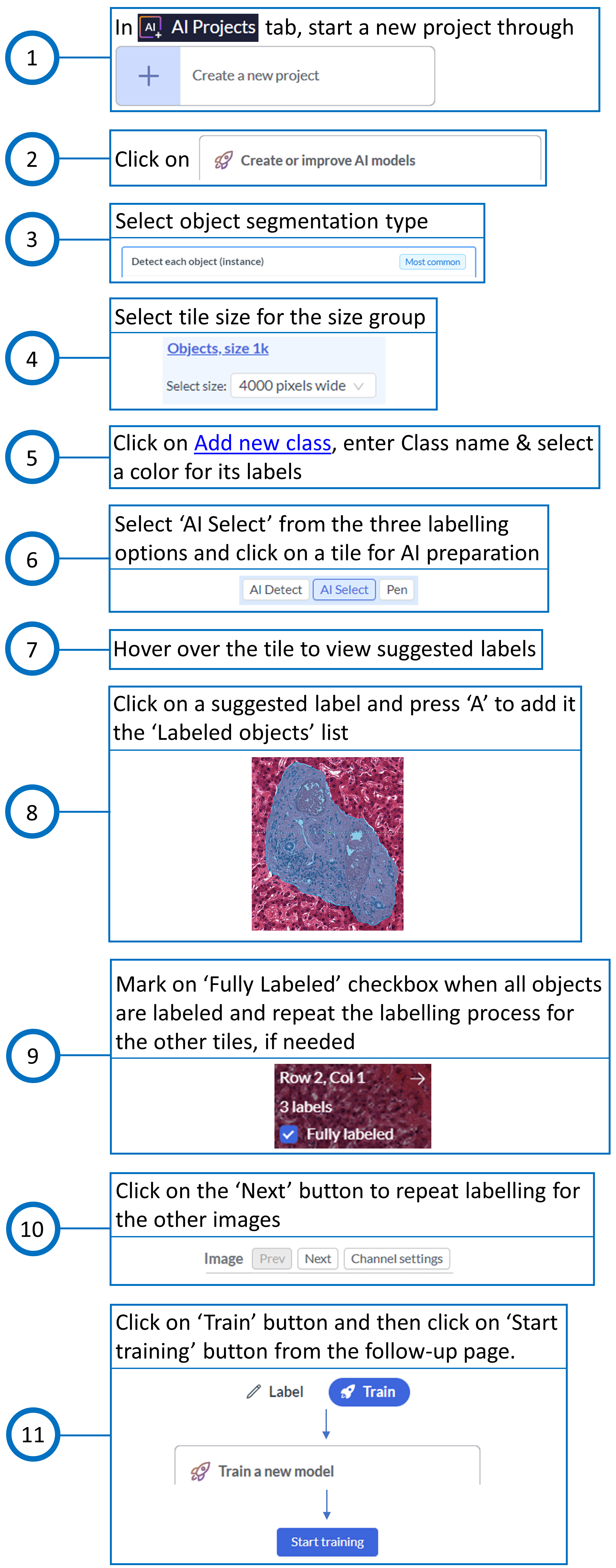
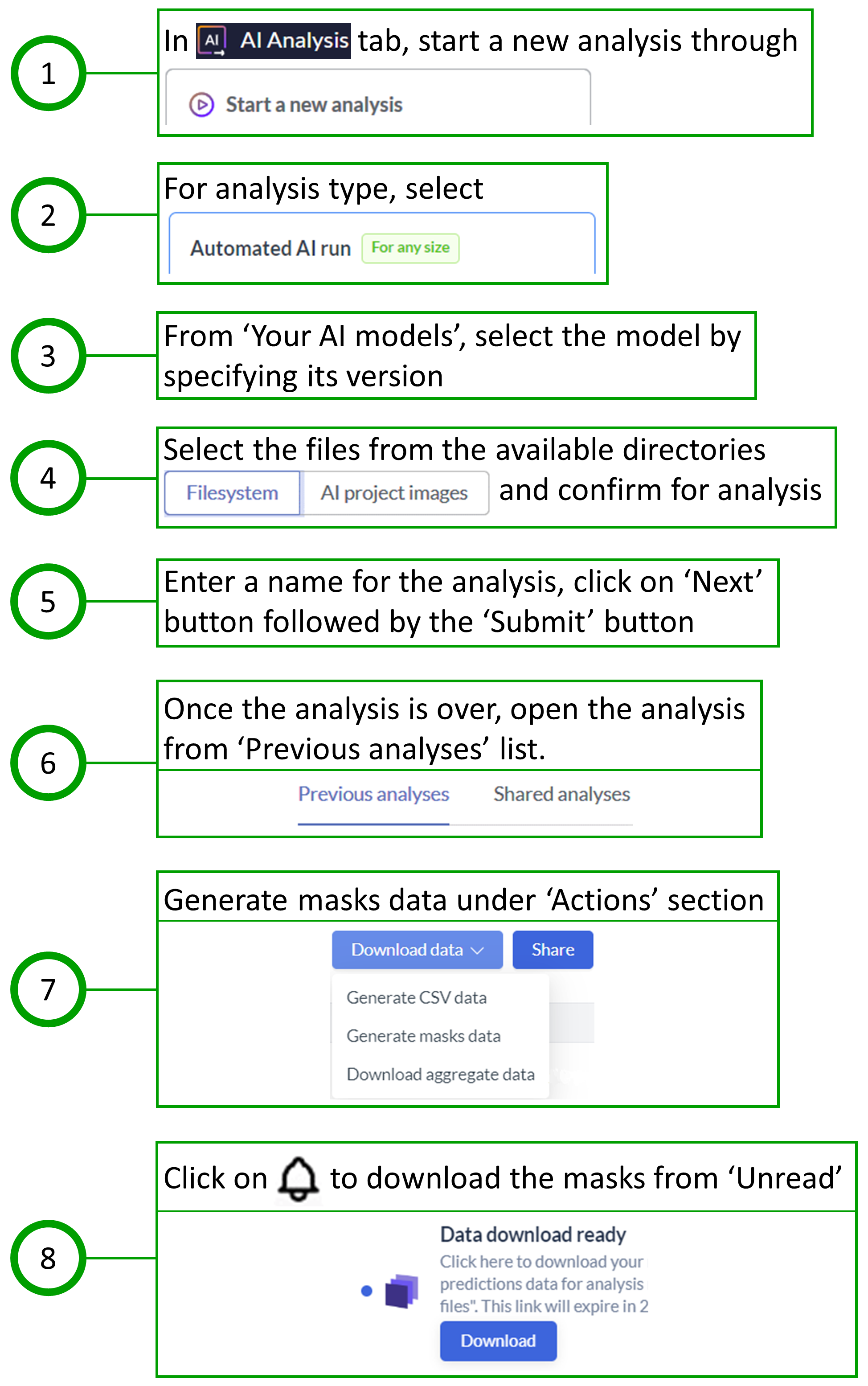
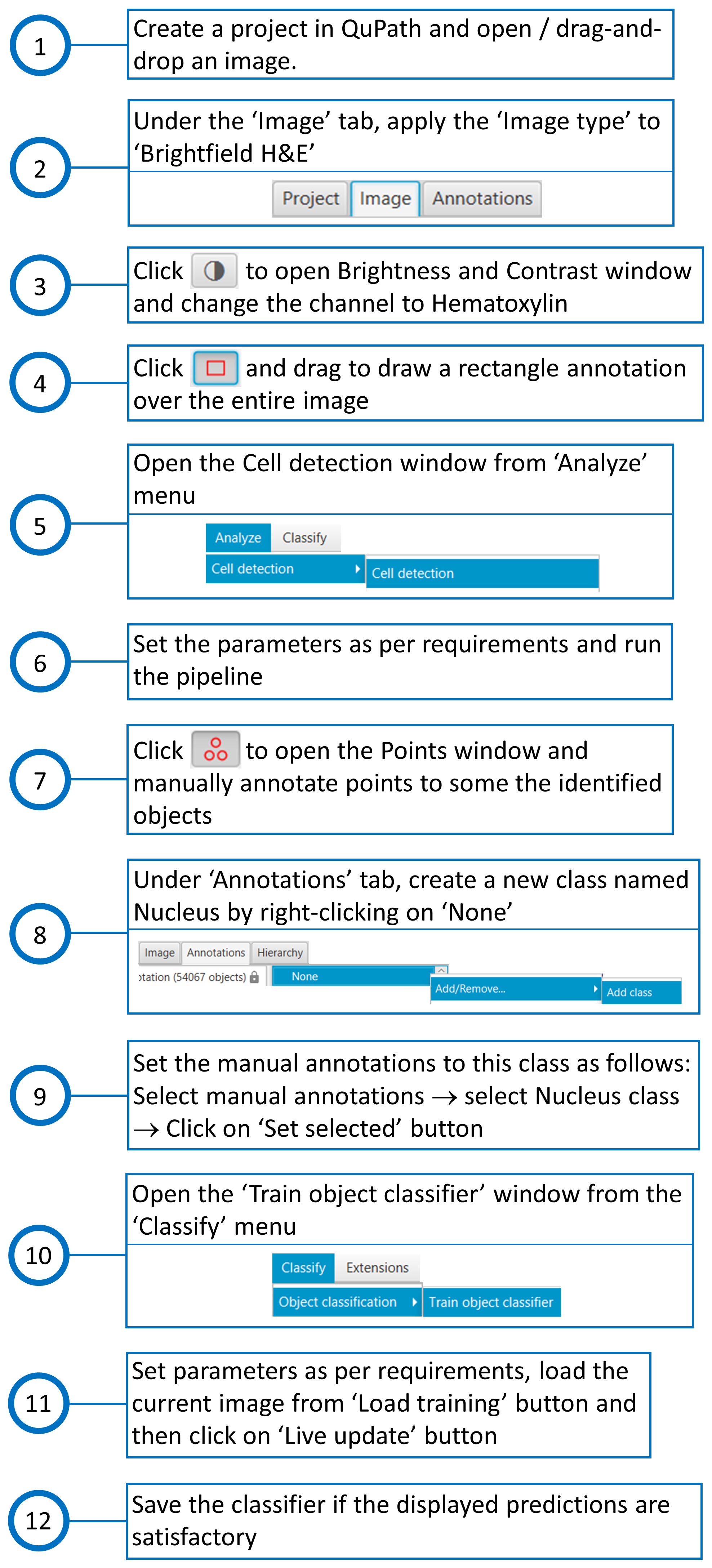
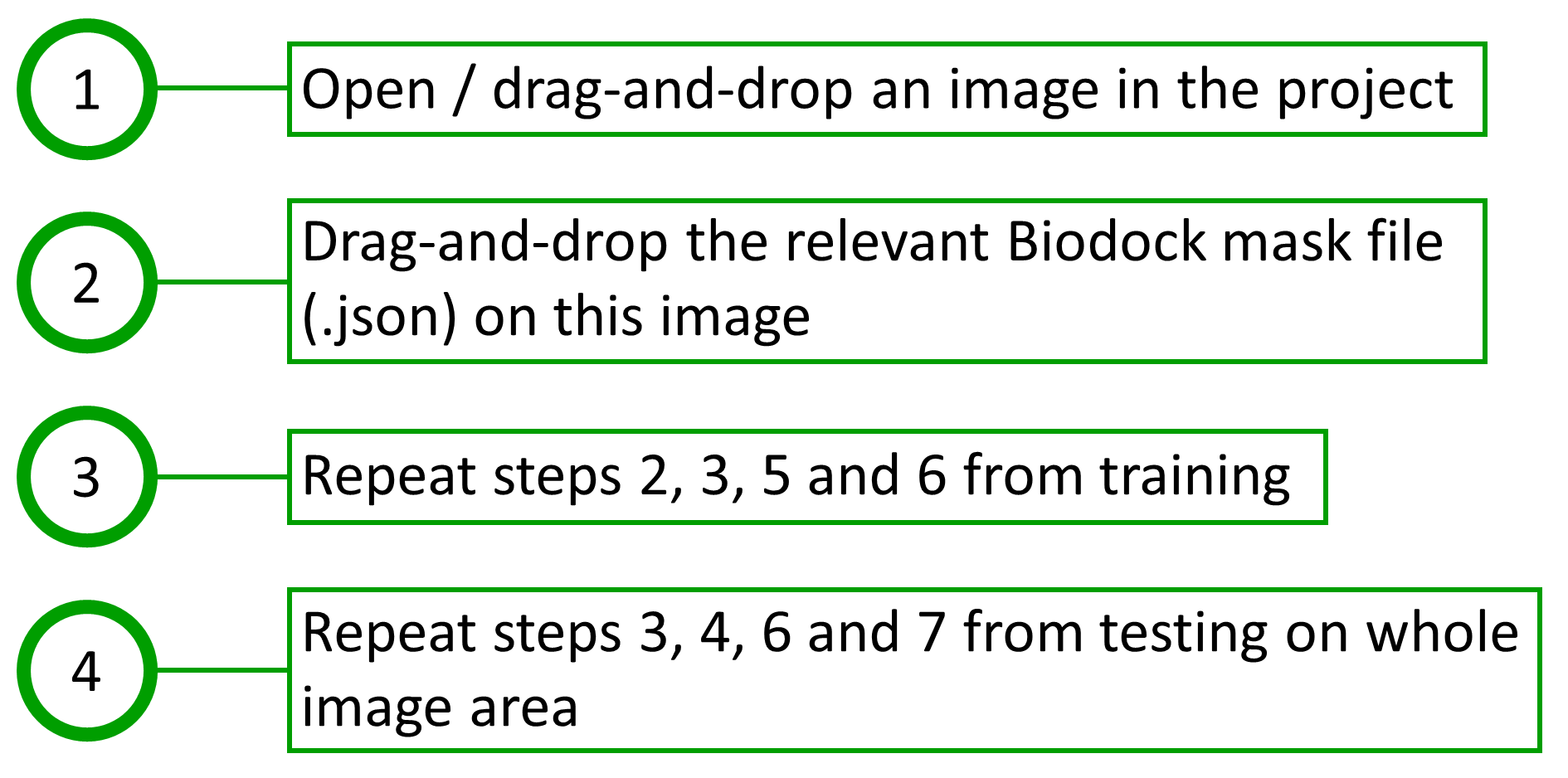
The steps taken for training in QuPath are shown in Fig. 5. The procedure for testing in QuPath was divided into two segments: one that identifies nuclei in the whole image area, and the other that identifies nuclei within the boundaries of Biodock masks, i.e., within each portal tract polygon. The steps taken for testing in QuPath are shown in Fig. 6 and Fig. 7. The measurements, when finally exported from QuPath, consist of a merged report on portal tracts and nuclei.

Conclusion
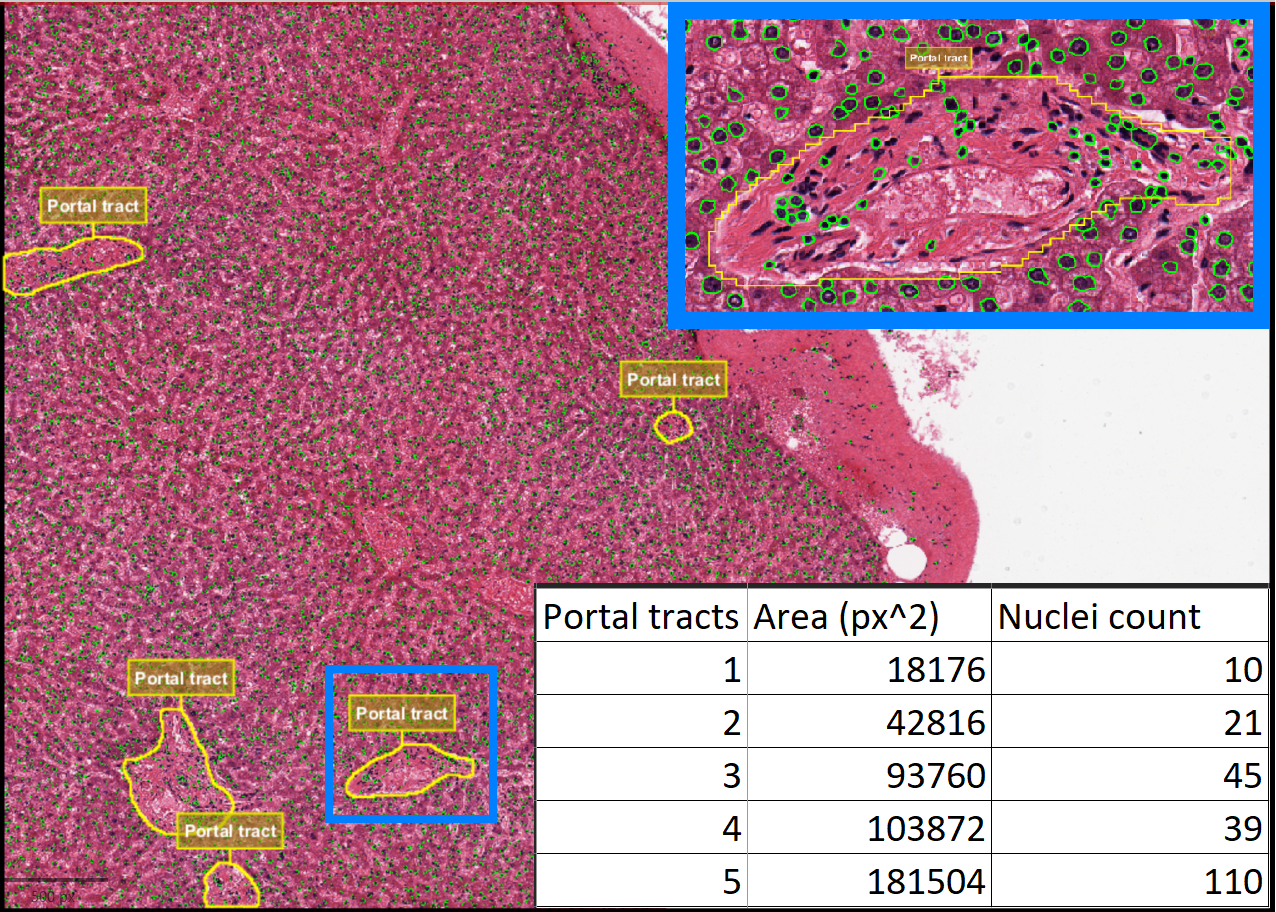
Based on the results from the test data, it was observed that fusing data from multiple tools can be highly beneficial for achieving the desired output, while also optimizing resource utilization. By combining the strengths of different tools, the process not only becomes more efficient but also allows for a more comprehensive analysis. The count of nuclei within each portal tract (Fig. 8) was successfully obtained. While QuPath was primarily used to detect nuclei in this instance, it has the potential to deliver more accurate results when it comes to identifying entire cells, particularly by leveraging its advanced cell detection feature, as discussed earlier.