Anyone with moderate to severe nearsightedness (myopia) will tell you that losing their glasses is devastating. I learned this the hard way in Costa Rica when the Pacific Ocean swallowed my pair, and I was left with my brutal myopia for the rest of the trip. I recall sitting at restaurants and looking confusedly at menus. I could make out where words were written but I did not know where one word started and another ended. Other times, when looking at people in the distance, I could make out the presence of a crowd but was unable to distinguish individuals and ascertain how many people were present. It was not until I looked closer at the menus and walked nearer to the crowds that individual words and people came into view. This silly albeit tragic tale evinces a key aspect of visual discrimination and classification: that is that there is a difference between distinguishing between two things and identifying the constitutive units of a thing among its group.
This discriminative ability has become a primary imperative of image segmentation models and can be distinguished by the names semantic and instance segmentation. Such models have also become useful in the field of biology where instance and semantic distinctions are made on histological images across many subfields. Here, I will discuss the differences between semantic and instance segmentation approaches for images of biological tissue by underscoring how extant models are being used to solve real biological problems with image analysis.
Table of contents:
1. Semantic vs. Instance segmentation: The basics
2. Semantic Segmentation
a. Continuous distinctions: Boundary Detection
b. Categorical distinctions: Disease detection
3. Instance Segmentation
a. Cell count and nuclear organization with Nuclei detection
b. Bacterial colonies
c. ROI extraction for Calcium Imaging
4. Semantic vs. instance segmentation: Which to choose and why?
Semantic segmentation finds the crowd whereas instance segmentation finds the individuals
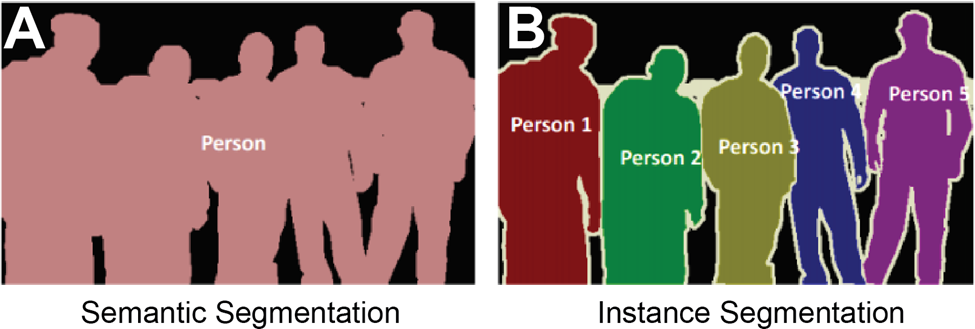
Consider a crowded room of your friends. In a snapshot of the image, semantic and instance segmentation models can be used to pick out people from the background. In the semantic model the output would be a silhouette of the whole group against the background (Figure 1a). In the instance model, the output would be the silhouettes of each person against each other and the background (Figure 1b). The fundamental reason behind these outputs is that each model is being asked to perform different tasks. The semantic model is often performing a binary classification for each pixel of the image, that is: is this pixel part of the person category or the background category? Meanwhile, the instance model is being asked to perform a binary classification and object distinction, that is: is this pixel part of a person category or the background category? And… which person does this pixel belong to?
Both questions have vital relevance in biology where images of tissues are routinely analyzed to distinguish qualities and individuals. What sorts of images are being analyzed and what classifications are such models being tasked with making?
Semantic Segmentation in Biological Samples
Semantic segmentation finds the boundaries of cells and multicellular structures to quantify continuous morphological features.
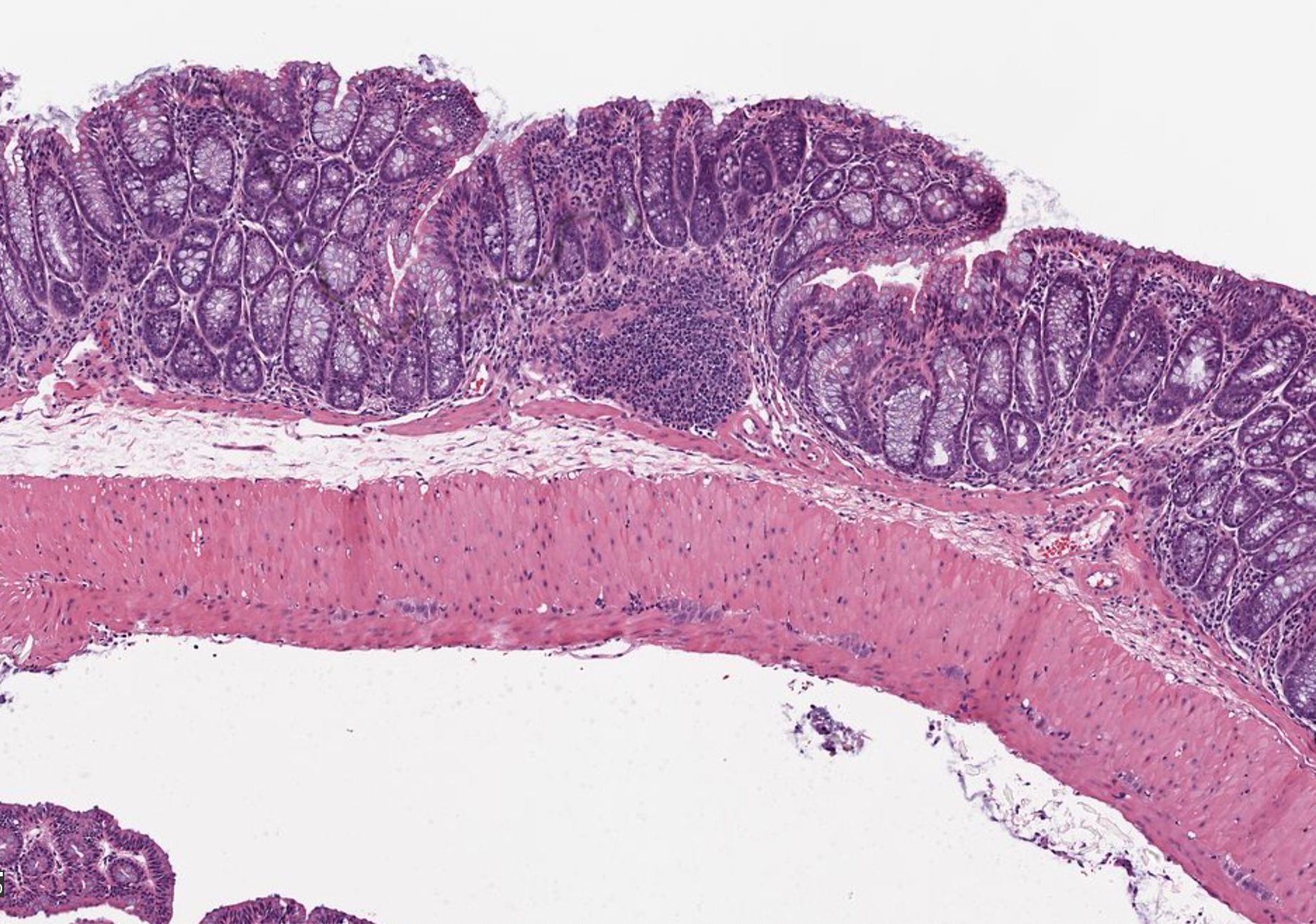
One powerful application for semantic models is the classification of cellular membranes against extracellular and intracellular space. This classification is performed routinely across fields and histological samples and is often used as a base to iterate upon for instance segmentation. At a basic level, a machine learning model is tasked with classifying pixels as being a member of a cellular boundary or not. The prediction can be read out in an image where each pixel value represents the confidence of the model in making that prediction (Figure 2).
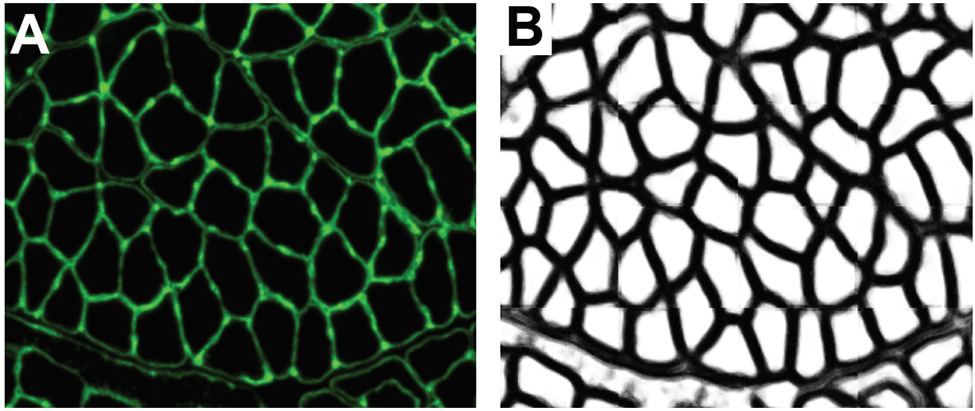
In muscle biology, a program I developed called Myosoft, performs this classification on sections of muscle tissue. The output of this classified image enables the program to ultimately perform instance segmentation to extract morphological information on individual muscle fibers across a section of the whole muscle. This allows you to quantify the distribution of muscle fiber morphology such as cross-sectional area which is a crucial metric influenced by physiological changes and disease states. Although this is an example of boundary detection applied to muscle tissue, similar models have been developed for adipose tissue, cerebral tissue, and plant cell walls.
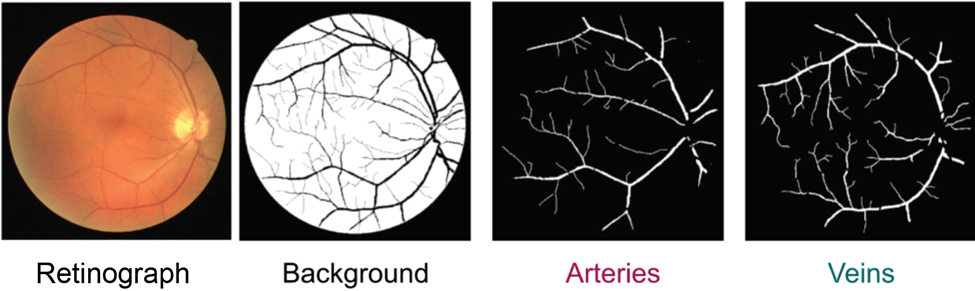
Semantic models are also predicting the boundaries of more complex multicellular structures such as veins. The lab of Jose Rouco in Spain developed a model to segment retinal arteries and veins. They implemented a convolutional neural network-based model to perform a binary classification on processed retinographs. The model outputs predictions for whether a pixel belongs to an artery/vein or extra-retinal space (vitreous humor) (Figure 3). From this prediction, scientists can extract relevant morphological features of the segmented vasculature such as length, branches, and surface area covered. This information can be usefully integrated into diagnostic approaches for retinal diseases such as Macular Degeneration, glaucoma, and diabetes all alter the architecture of retinal vasculature.
Semantic Segmentation makes categorical classifications to automatically identify distinct regions and disease states
Although semantic segmentation can be used to define ROIs that can be quantified to extract continuous variables such as cross-sectional area, it also has widespread potential for making categorical classifications. In the field of biology, this is especially useful for identifying disease states and histological features/regions of interest.
Sickle cell anemia is caused by a point mutation which results in red blood cells assuming a deformed sickle shape which makes them inefficient oxygen carriers. Due to the distinct morphological profile of sickle cell red blood cells, one group developed a deep learning-based model to perform semantic segmentation on red blood cells where the model would find red blood cells and then classify them as either sickled or healthy. They demonstrated that their model could predict red blood cell type with high accuracy. Incorporating multiple classes of red blood cells beyond sickle vs healthy was also successful. This presents potential as a powerful diagnostic tool and demonstrates how semantic segmentation can be applied to discriminate disease states when such states are associated with morphological differences. Beyond discriminating cell types to distinguish a disease state, semantic models can also be used to classify images of whole tissue and identify regions of interest.
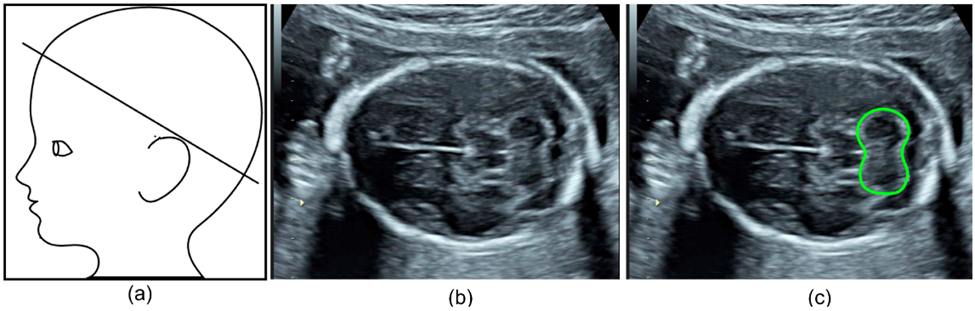
Specifically, one group aimed to automate the detection and segmentation of the fetal cerebellum. The cerebellum is a brain region comprised of a dense network of neurons involved in motor coordination and motion calibration. This structure is also used as a benchmark for brain development during fetal ultrasound scans as structural changes report neurodevelopmental stages and might report neurodevelopmental outcomes. The authors developed a deep learning algorithm to automatically segment the cerebellum in two-dimensional ultrasound images (Figure 4b). Their model was highly accurate with an 86% overlap compared to manual annotation, making it a reliable and efficient alternative.
Instance segmentation in Biological Samples
Instance segmentation reliably reveals the distribution, quantity, and structural profile of individual cells and cellular components over space.
One nearly ubiquitously imaged and analyzed cellular feature is the nucleus. Not only does the nucleus offer a direct measure of cell count and location but it is also used as a biomarker for certain cellular states. For example, In muscle fibers, translocation of the nucleus to the center of the fiber, termed centrally nucleated myofibers, is a biomarker for damage and typically occurs following muscle stress. This analysis, howsoever important, is typically performed manually which is labor-intensive and unreliable in large sections that can contain thousands of nuclei. Therefore, many groups have aimed to develop automated ways of segmenting individual nuclei.
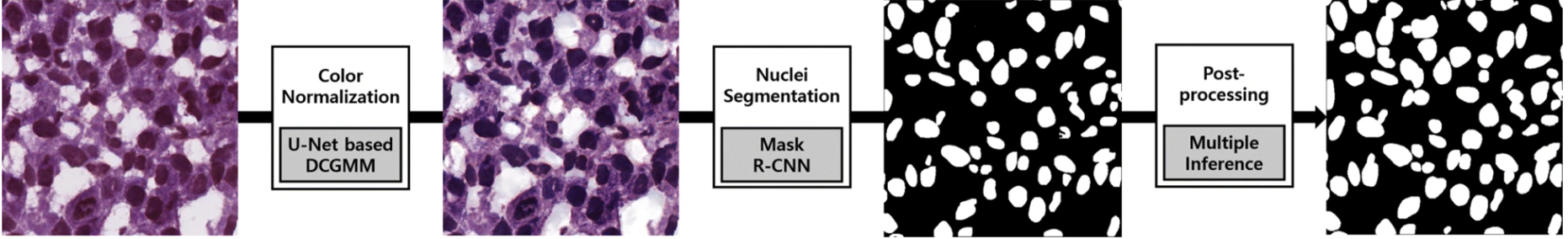
This is a prime task for an instance segmentation model. One group used a Mask R-convolutional neural network as their segmentation framework to develop a deep learning-based model to segment nuclei in H&E-stained images (Figure 5). What’s impressive about their program is that it was able to perform accurate instance segmentation of nuclei over a variety of tissue types such as liver, prostate, and colon. Furthermore, accuracy was consistent across patients. This demonstrates the model’s ability to extract instances of nuclei in cases of image variability.
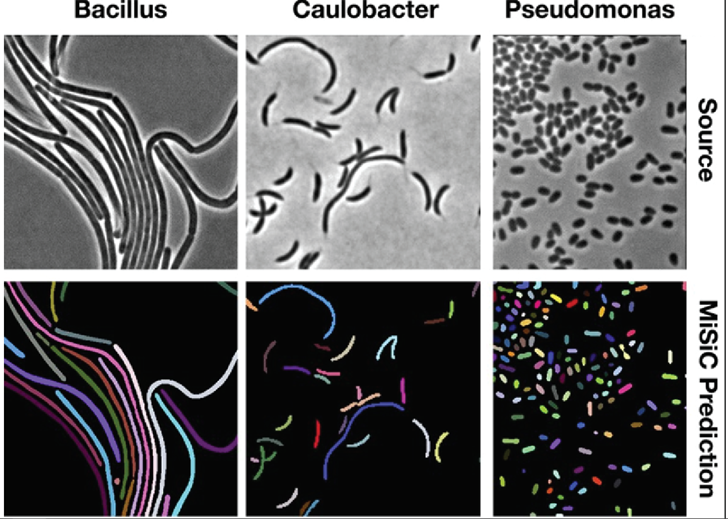
Models for instance segmentation can also go beyond identifying homogenous cellular features but can be generalized to segmenting whole individual cells. In one such example, a group developed a U-Net based framework to perform instance segmentation of bacteria. Quantifying the distribution and morphology of bacterial colonies is an essential metric for understanding bacterial growth and microbiome diversity. In their study, they were able to apply their network to segment different strains of bacteria over different microscopy techniques (Figure 6).
These examples demonstrate the application of instance segmentation to define the quantity and distribution of cellular components over space. How, though, might instance segmentation be used to define the distribution of cells to ultimately determine their physiological state over time?
Utilizing instance segmentation to define cell location and physiological state over time
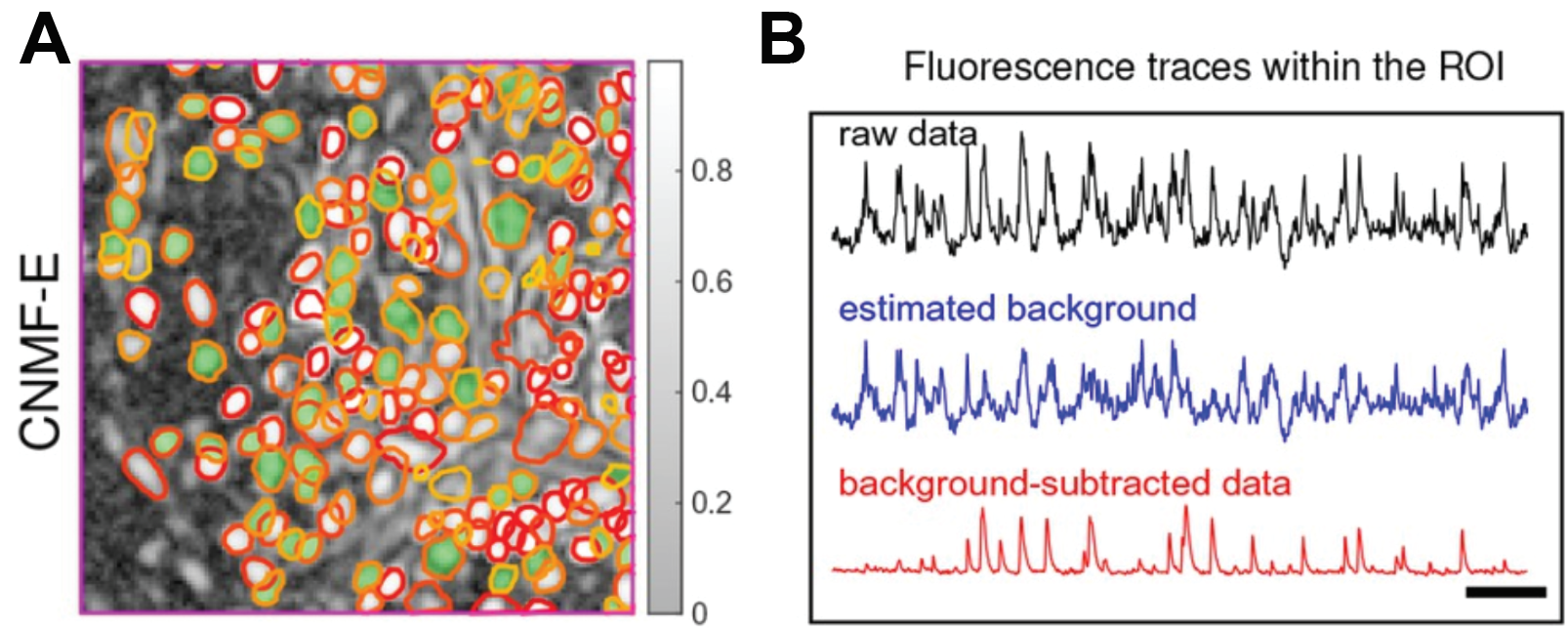
In neuroscience, optical readouts of neuronal activity have become ubiquitous tools for understanding neural circuit function. This method fundamentally works by impregnating neurons with a protein that reports calcium binding with light. In this way, the concentration of calcium can be read out optically since calcium is directly tied to neural activity. Neural activity can be read out in turn. For more information on calcium imaging, see my article on holographic optogenetics! Obtaining the activity of individual neurons necessitates defining where that neuron is located in space and extracting the light intensity of that region over time. Traditionally this was performed manually which is tedious and does not permit the demixing of neural activity of overlapping cells. Recently, however, a group implemented constrained non-negative matrix factorization, an unsupervised learning algorithm, to perform instance segmentation on raw imaging data. This segments individual neurons within the population and allows for the extraction of intensity from individual cells (Figure 7).
The advantages and disadvantages of semantic and instance segmentation are complementary
Both semantic and instance segmentation have their drawbacks and advantages. For semantic segmentation, the primary advantage is that it is easily implementable and highly accurate. Since the model is performing binary classification, additional steps to segment instances do not have to be taken into consideration. As a direct effect, however, this limits your analysis to quantifying qualities of the whole morphology of what you are studying. Conversely, Instance segmentation suffers from being harder to implement and achieve high accuracy from. As a result, many methods depend on some user supervision to revise and check the automated output. The benefit, however, is that with access to individual units, the dimensionality of data is much higher. The decision between implementing semantic or instance segmentation depends on the nature of the question you are asking as a scientist. Are you interested in characterizing the qualities of one structure or many structures? Do you need individual data or will population data suffice to address your hypothesis? With whichever segmentation framework you chose, you can use Biodock to design a custom AI module to meet your needs! Reach out to examine all the services Biodock has to offer.