Improving the resolution of optical microscopes was thought to be limited by a theorem put forward in the 19th century. However, scientists have been able to overcome this issue in the 21stcentury.
In this piece we’ll go over:
-A recent work that improves on our current super resolution technologies.
-The biological findings.
-The importance of microscopy in the field of immunology and image analysis.
In the 16-17th century Europe, a Dutch tradesman and scientist named Antoine van Leeuwenhoek used microscopes to discover red blood cells and to first see living sperm cells of animals. In 1873, there was an equation published by Ernst Abbe demonstrating how the resolution of a light microscope was limited by, among other things, the wavelength of light. Through the 19th and much of the 20th century this limit, Abbe’s Limit (0.2 um), was thought to be unsurpassable. However, in 2014, the Nobel Prize in Chemistry was awarded for a technique called single-molecule localization microscopy (SMLM) which bypasses Abbe’s diffraction limit and set a new precedent for the field of microscopy—resolution at the nanoscale level. Today, SMLM approaches have the potential to capture natural processes at the nanoscale level such as molecular complexes, protein-protein interactions, and spatial organizations.
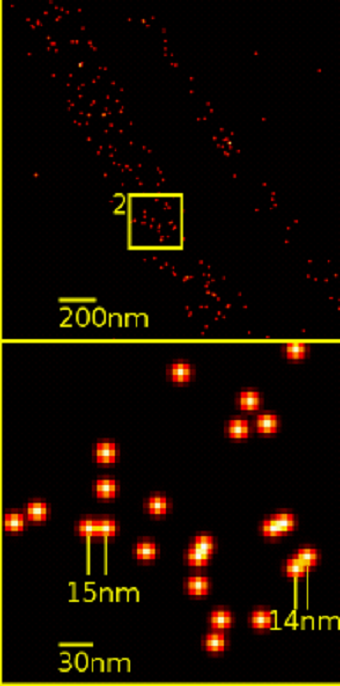
A recent work by Coelho et al. leverages this technique, improves upon it, and uses it to look closely at an immune cell called a T cell. The two primary super resolution techniques are stochastic optical reconstruction microscopy (STORM) and DNA point accumulation for imaging in nanoscale topography (DNA-PAINT). In SMLM, single fluorophores are temporally separated by using a weak light pulse to activate a fraction of all the fluorophores. These fluorophores glow until bleached at which point the process is repeated for a new set of fluorophores. Next, the blurry images are mathematically processed using probability theory (i.e. where is the original point source of the light?) to render them much sharper into points. Eventually every frame is superimposed to form one final, super resolution image. However, the time required to sequential image all the individual fluorophores require long acquisition times, as typically tens of thousands of frames are needed to map a given protein species in a cell. Drift during camera exposure affects the localization precision and the accuracy of localization. Thus, in practice the resolution is reduced to tens of nanometers making it unfeasible to conduct distance measurements on biological relevant scales.
Coelho et al. overcome this issue using an engineering solution coined Feedback SMLM, “…which can capture molecular emissions in a complex and unknown cellular environment with equal probability and high precision.” In other words, their technique can measure distances between two proteins in a wide range of sample formats, including intact cells, to capture heterogeneity as well as rare events with nanometer resolution.
Feedback SMLM uses three types of corrections: stage-sample feedback loop, autonomous optical feedback loop and piezoelectric mirror correction. Stage-sample feedback is accomplished by using non-fluorescent fiducials (reference points) outside of the field of view. A camera operating at a speed of 370 frames per second tracks the fiducials to provide information of both x-y and z position. Stage corrections can happen during sample acquisition with stabilization of 0.4 and 1 nm (SD) in the lateral (x-y) and axial (z) directions, respectively, over hours and days. Optical feedback uses a white LED integrated in the microscope body, creating an optical fiducial, which a camera is monitoring with a precision of 0.05 nm. Finally, the piezoelectric mirror tracks the mechanical instabilities and reduces image drift to 0.22 nm. The mirror correction accomplishes something other SMLM techniques can not; that is the stability of fluorescence path does not have to be computationally corrected post-acquisition.
Using DNA-PAINT with Feedback SMLM the authors generate an ultrahigh resolution image of F-actin. F-actin is an abundant protein in cells and used as an imaging benchmark because the width of actin is sub diffraction limit (i.e <0.2 um). Incredibly an individual actin fibril can be accurately measured to width of 5 to 9 nm without any post-acquisition processing. Electron microscopy measurements corroborate these results. In contrast other current SMLM imaging approaches, e.g. dSTORM, were only able to obtain an approximate width of 20 nm due to insufficient localization precision.
Finally, the authors apply their technique to an unanswered immunological question: how do T cells (derived from the Thymus) signal to become activated? T cells activate through their T cell receptor (TCR) which recognizes peptides presented in major histocompatibility complex (pMHC). T cells are exquistively sensitive and can recognize fewer than 10 agonist pMHC ligands with only a few TCRs. The current model suggests a phosphorylation cascade on associated protein dimers (i.e. CD3 dimers) of the TCR triggers cellular activation. It is widely assumed TCR triggering requires the exclusion of the transmembrane phosphatase CD45 to successfully activate. However, these delicate events have never been visualized before and are currently readout biochemically. Coelho et al. activated Jurkat T cells using pMHC coated on a lipid bilayer glass slide. The authors applied Feedback SMLM and DNA-PAINT to monitor CD45 and phosphorylated CD3ζ (pCD3ζ). What they found was spatially separated nano-clusters of pCD3ζ and CD45 at median distances of 19.6 nm. On resting T cells CD3 and CD45 appeared intermix with mean distances of 12.5 nm. Thus, if spatial separation was the means of TCR triggering, the TCR is exquisitely sensitive to 4- to 7-nm differences between CD3 and CD45. Through careful microscopy image analysis, the author's feedback SMLM has provided the means to answer a question previously unknown due to technological reasons and provides new insights into the field of T cell biology.
Reference
1) Coelho et al. Ultraprecise single-molecule localization microscopy enables in situ distance measurements in intact cells. Science Advances. Vol 6, Issue 16. 2020. DOI: 10.1126/sciadv.aay8271


